aslprep.sdcflows.workflows.pepolar module
Datasets with multiple phase encoded directions.
PEPOLAR techniques
This corresponds to this section of the BIDS specification.
- init_pepolar_unwarp_wf(omp_nthreads=1, matched_pe=False, name='pepolar_unwarp_wf')[source]
Create the PE-Polar field estimation workflow.
This workflow takes in a set of EPI files with opposite phase encoding direction than the target file and calculates a displacements field (in other words, an ANTs-compatible warp file).
This procedure works if there is only one
_epifile is present (as long as it has the opposite phase encoding direction to the target file). The target file will be used to estimate the field distortion. However, if there is another_epifile present with a matching phase encoding direction to the target it will be used instead.Currently, different phase encoding directions in the target file and the
_epifile(s) (for example,iandj) is not supported.The warp field correcting for the distortions is estimated using AFNI’s
3dQwarp, with displacement estimation limited to the target file phase encoding direction.It also calculates a new mask for the input dataset that takes into account the distortions.
- Workflow Graph
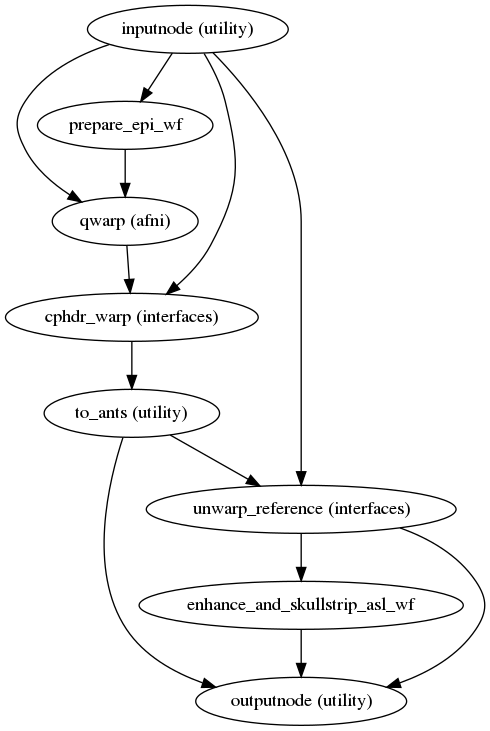
(Source code, png, svg, pdf)
- Parameters:
matched_pe (bool) – Whether the input
fmaps_epiwill contain images with matched PE blips or not. Please usesdcflows.workflows.pepolar.check_pes()to determine whether they exist or not.name (str) – Name for this workflow
omp_nthreads (int) – Parallelize internal tasks across the number of CPUs given by this option.
- Inputs:
fmaps_epi (list of tuple(pathlike, str)) – The list of EPI images that will be used in PE-Polar correction, and their corresponding
PhaseEncodingDirectionmetadata. The workflow will use theepi_pe_dirinput to separate out those EPI acquisitions with opposed PE blips and those with matched PE blips (the latter could be none, andin_reference_brainwould then be used). The workflow raises aValueErrorwhen no images with opposed PE blips are found.epi_pe_dir (str) – The baseline PE direction.
in_reference (pathlike) – The baseline reference image (must correspond to
epi_pe_dir).in_reference_brain (pathlike) – The reference image above, but skullstripped.
in_mask (pathlike) – Not used, present only for consistency across fieldmap estimation workflows.
- Outputs:
out_reference (pathlike) – The
in_referenceafter unwarpingout_reference_brain (pathlike) – The
in_referenceafter unwarping and skullstrippingout_warp (pathlike) – The corresponding DFM compatible with ANTs.
out_mask (pathlike) – Mask of the unwarped input file
- init_prepare_epi_wf(omp_nthreads, matched_pe=False, name='prepare_epi_wf')[source]
Prepare opposed-PE EPI images for PE-POLAR SDC.
This workflow takes in a set of EPI files and returns two 3D volumes with matching and opposed PE directions, ready to be used in field distortion estimation.
The procedure involves: estimating a robust template using FreeSurfer’s
mri_robust_template, bias field correction using ANTsN4BiasFieldCorrectionand AFNI3dUnifize, skullstripping using FSL BET and AFNI3dAutomask, and rigid coregistration to the reference using ANTs.- Workflow Graph
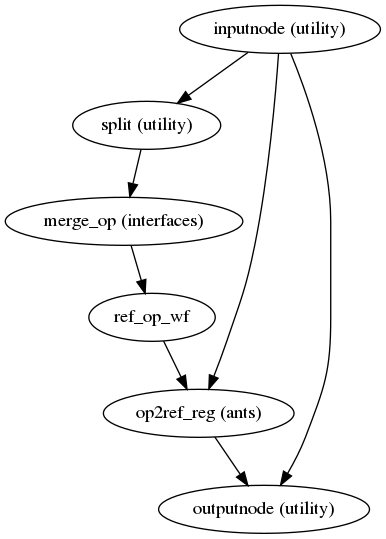
(Source code, png, svg, pdf)
- Parameters:
matched_pe (bool) – Whether the input
fmaps_epiwill contain images with matched PE blips or not. Please usesdcflows.workflows.pepolar.check_pes()to determine whether they exist or not.name (str) – Name for this workflow
omp_nthreads (int) – Parallelize internal tasks across the number of CPUs given by this option.
- Inputs:
epi_pe (str) – Phase-encoding direction of the EPI image to be corrected.
maps_pe (list of tuple(pathlike, str)) – list of 3D or 4D NIfTI images
ref_brain – coregistration reference (skullstripped and bias field corrected)
- Outputs:
opposed_pe (pathlike) – single 3D NIfTI file
matched_pe (pathlike) – single 3D NIfTI file