aslprep.workflows.asl.base module
Preprocessing workflows for ASL data.
- init_asl_wf(*, asl_file: str, precomputed: dict = {}, fieldmap_id: str | None = None)[source]
Perform the functional preprocessing stages of ASLPrep.
- Workflow Graph
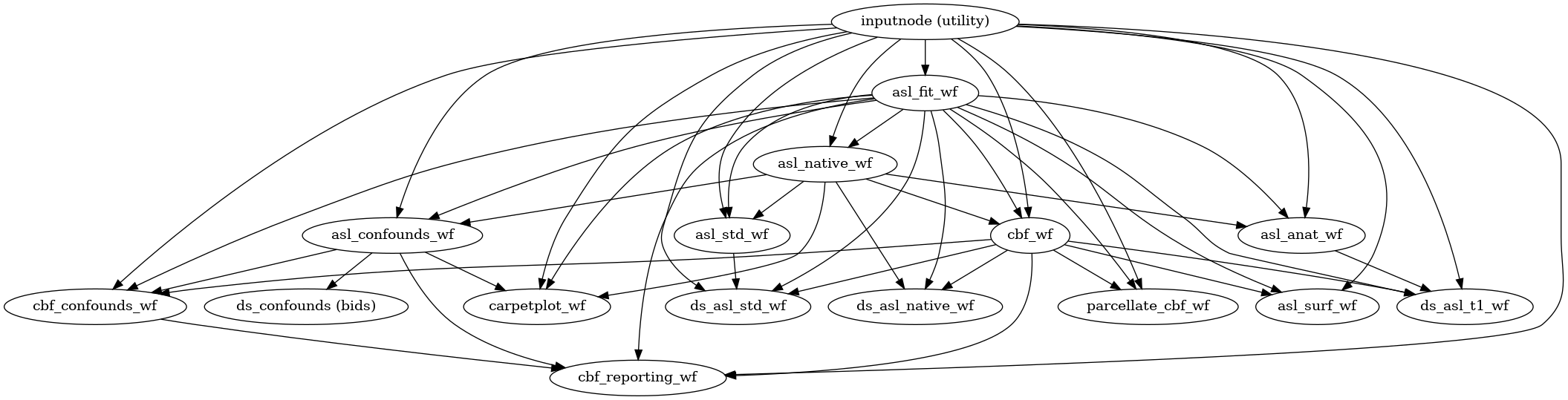
(Source code, png, svg, pdf)
- Parameters:
asl_file – ASL NIfTI file
precomputed – Dictionary containing precomputed derivatives to reuse, if possible.
fieldmap_id – ID of the fieldmap to use to correct this BOLD series. If
None, no correction will be applied.
- Inputs:
asl_file – asl series NIfTI file
t1w_preproc – Bias-corrected structural template image
t1w_mask – Mask of the skull-stripped template image
t1w_dseg – Segmentation of preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF)
t1w_tpms – List of tissue probability maps in T1w space
fmap_id – Unique identifiers to select fieldmap files
fmap – List of estimated fieldmaps (collated with fmap_id)
fmap_ref – List of fieldmap reference files (collated with fmap_id)
fmap_coeff – List of lists of spline coefficient files (collated with fmap_id)
fmap_mask – List of fieldmap masks (collated with fmap_id)
sdc_method – List of fieldmap correction method names (collated with fmap_id)
template – List of templates to target
anat2std_xfm – List of transform files, collated with templates
- Outputs:
asl_t1 – asl series, resampled to T1w space
asl_mask_t1 – asl series mask in T1w space
asl_std – asl series, resampled to template space
asl_mask_std – asl series mask in template space
confounds – TSV of confounds
cbf_ts_t1 – cbf times series in T1w space
mean_cbf_t1 – mean cbf in T1w space
cbf_ts_score_t1 – scorecbf times series in T1w space
mean_cbf_score_t1 – mean score cbf in T1w space
mean_cbf_scrub_t1, mean_cbf_gm_basil_t1, mean_cbf_basil_t1 – scrub, parital volume corrected and basil cbf in T1w space
cbf_ts_std – cbf times series in template space
mean_cbf_std – mean cbf in template space
cbf_ts_score_std – scorecbf times series in template space
mean_cbf_score_std – mean score cbf in template space
mean_cbf_scrub_std, mean_cbf_gm_basil_std, mean_cbf_basil_std – scrub, parital volume corrected and basil cbf in template space
qc_file – quality control measures
Notes
Note that
anat2std_xfm,std_space,std_resolution,std_cohort,std_t1wandstd_maskare treated as single inputs. In order to resample to multiple target spaces, connect these fields to an iterable.Brain-mask T1w.
Generate initial ASL reference image. I think this is just the BOLD reference workflow from niworkflows, without any modifications for ASL data. I could be missing something. - Not in GE workflow. - In the GE workflow, the reference image comes from the M0 scan.
Motion correction. - Outputs the HMC transform and the motion parameters. - Not in GE workflow.
Susceptibility distortion correction. - Outputs the SDC transform. - Not in GE workflow.
Register ASL to T1w.
Apply the ASL-to-T1w transforms to get T1w-space outputs (passed along to derivatives workflow).
Apply the ASL-to-ASLref transforms to get native-space outputs. - Not in GE workflow.
Calculate confounds. - Not in GE workflow.
Calculate CBF.
Refine the brain mask.
Generate distortion correction report. - Not in GE workflow.
Apply ASL-to-template transforms to get template-space outputs.
CBF QC workflow.
CBF plotting workflow.
Generate carpet plots. - Not in GE workflow.
Parcellate CBF results.